Web Service Tutorial
The functions of the web service:
Overview
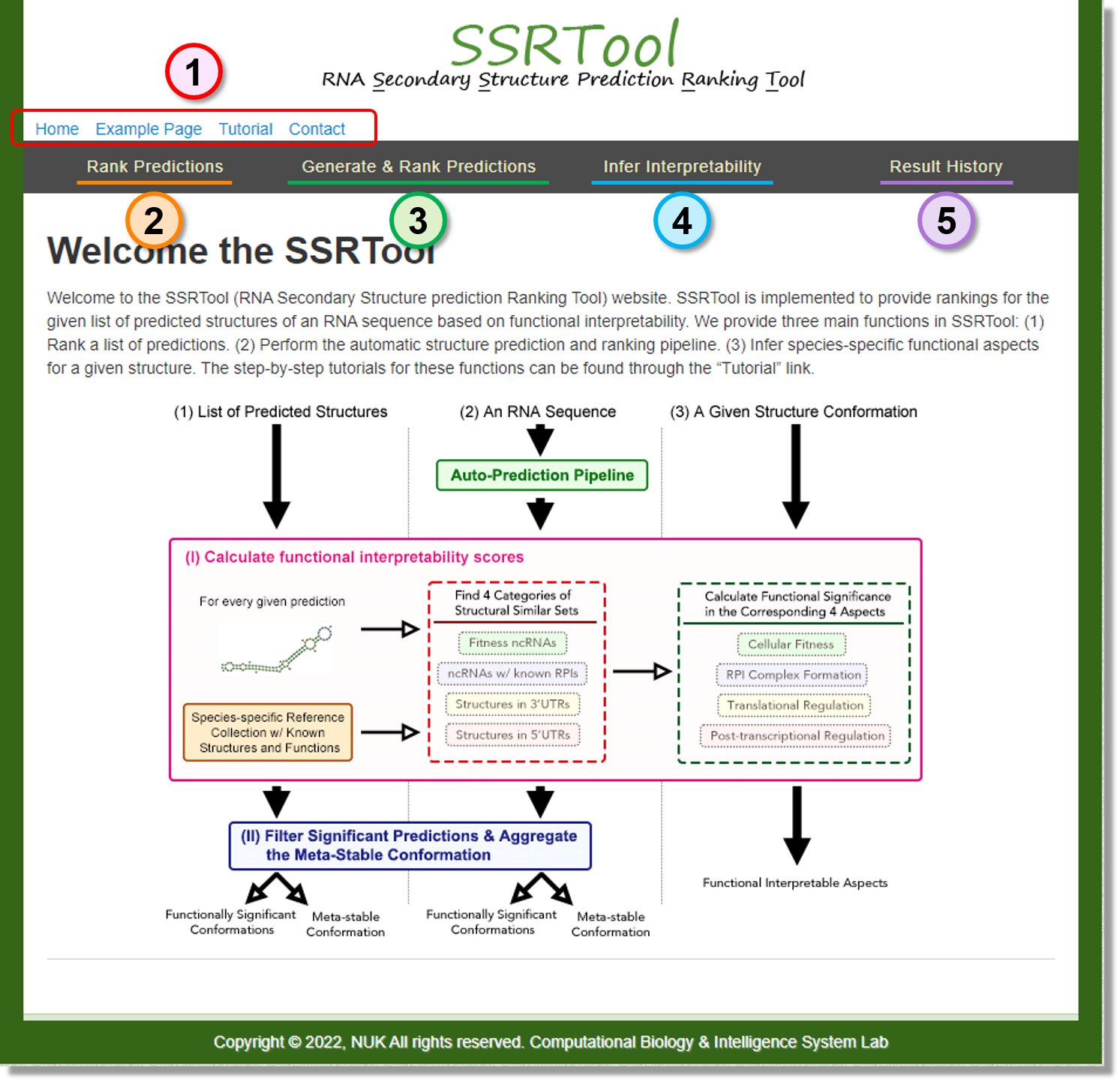
-
Home
Back to the "Home" page.
Example Page
We provide a sample result generated by the "Generate & Rank Predictions" function.
Tutorial
This page, users can learn how to use this service.
Contact
The information about us and the term of use. -
Rank Predictions
Rank the List of Predicted Structures Based on Functional Interpretability -
Generate & Rank Predictions
Generate the Functionally Interpretable Predictions for a Given Sequence -
Infer Interpretability
Infer the Functional Interpretability of a Given Structure -
Result History
Help users to use job_id to find their previously submitted results.
Rank Predictions
In this page, users can upload a .zip file of .ct files to rank the list of predicted sructures based on functional interpretability.
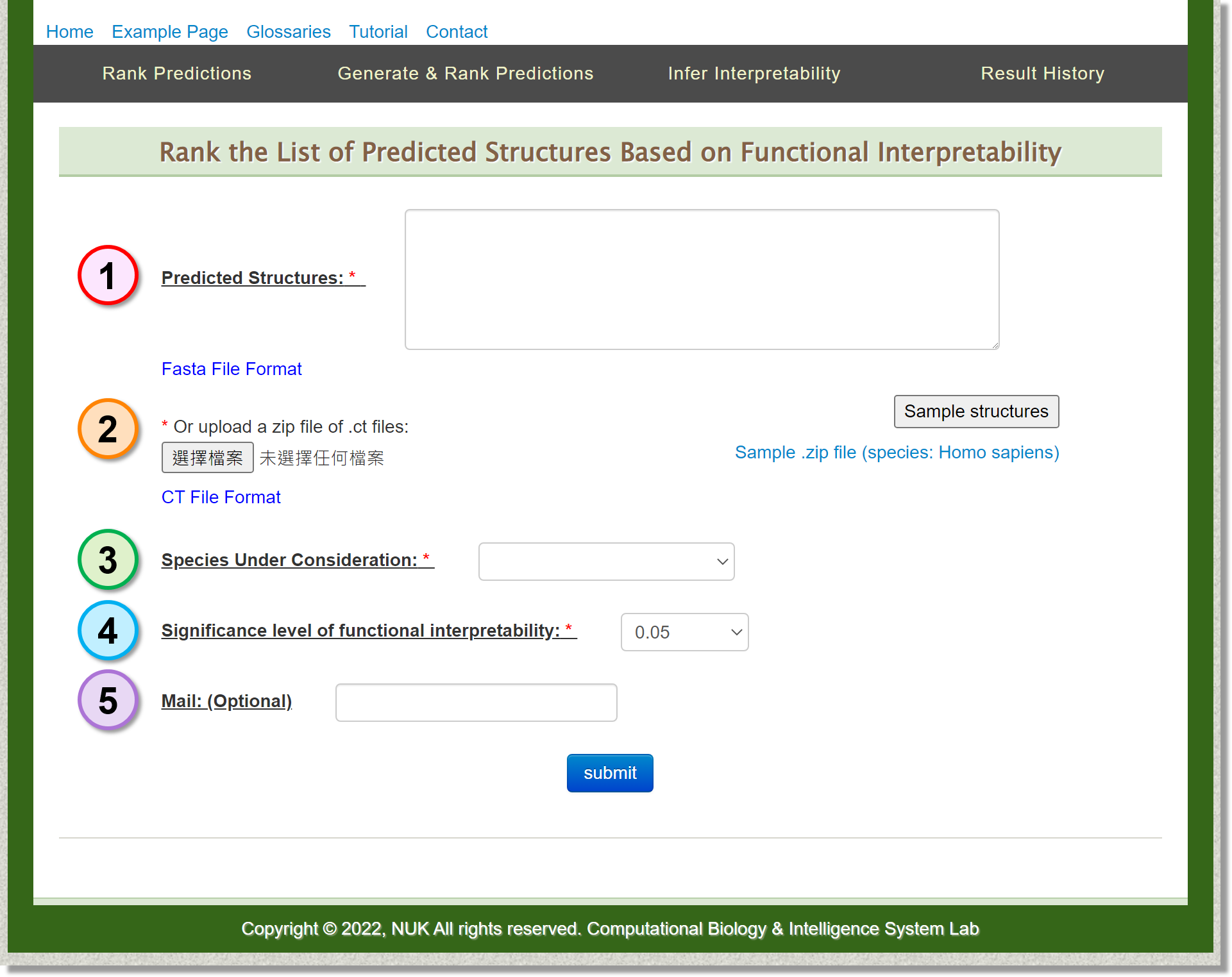
-
Input: Predicted Structures
Users can provide a list of structures in the fasta format. -
Input: Upload
Users can also upload your .zip file of .ct files as the input structures. -
Input: Species Under Consideration
Since the implemented prediction ranking algorithm considers species-specific functionality, targeted species should be chosen. In the current version, SSRTool provide the species of Arabidopsis, human, mouse, rat, yeast and zebrafish for users. -
Input: Significance level of functional interpretability
Set the significance level of functional interpretability. -
Input: Mail (Optional)
Depending on the workload of the server, it may take several minutes perform the job.
Users can optionally provide their mail addresses. A notification letter will be sent when the job is ready.
Generate & Rank Predictions
In this page, users can input the RNA sequence of interest for structure prediction and ranking.
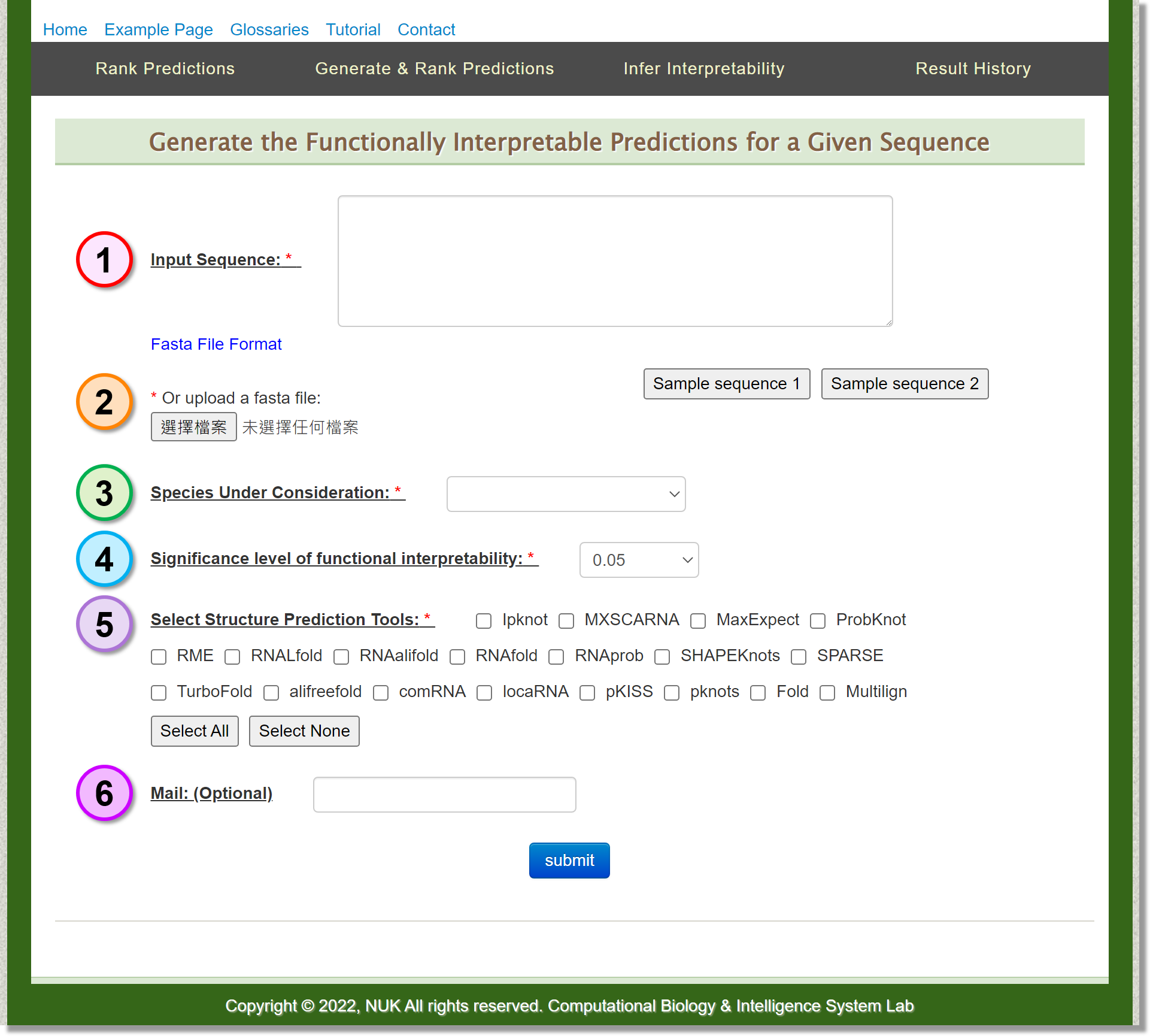
-
Input: Sequence
Users can provide the sequence of interest in .fasta format.
-
Input: Upload
Users can also upload the existing .fasta file. -
Input: Species
Since the implemented prediction ranking algorithm considers species-specific functionality, targeted species should be chosen. In the current version, SSRTool provide the species of Arabidopsis, human, mouse, rat, yeast and zebrafish for users. -
Input: Significance level of functional interpretability
Set the significance level of functional interpretability. -
Input: Select Structure Prediction Tools
Select the structure prediction tools to be used in the prediction process. -
Input: Mail (Optional)
Depending on the workload of the server, it may take several minutes perform the job.
Users can optionally provide their mail addresses. A notification letter will be sent when the job is ready.
Sample Sequence
Two sample sequences are provided for exploring the web service.
Infer Interpretability
Users can input the RNA structure or upload the .ct file to infer the functional interpretability of it.
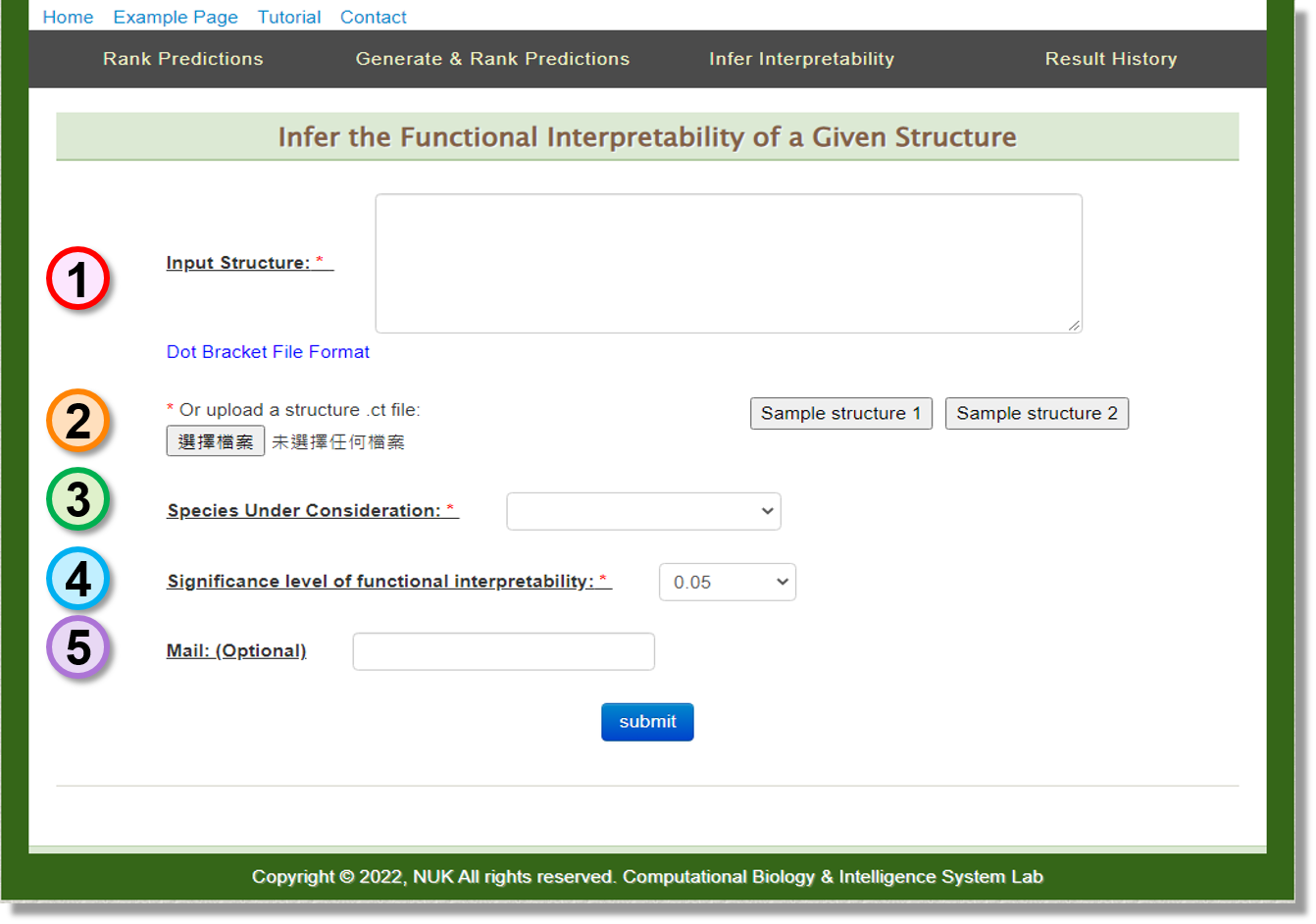
-
Input: Structure
Users can provide the structure of interest in the Dot Bracket File format. -
Input: Upload
Users can also upload the structure .ct file as the input structure. -
Input: Species Under Consideration
Since the implemented prediction ranking algorithm considers species-specific functionality, targeted species should be chosen. In the current version, SSRTool provide the species of Arabidopsis, human, mouse, rat, yeast and zebrafish for users. -
Input: Significance level of functional interpretability
Set the significance level of functional interpretability. -
Input: Mail (Optional)
Depending on the workload of the server, it may take several minutes perform the job.
Users can optionally provide their mail addresses. A notification letter will be sent when the job is ready.
Sample Sequence
Two sample structures are provided for exploring the web service.
Result History
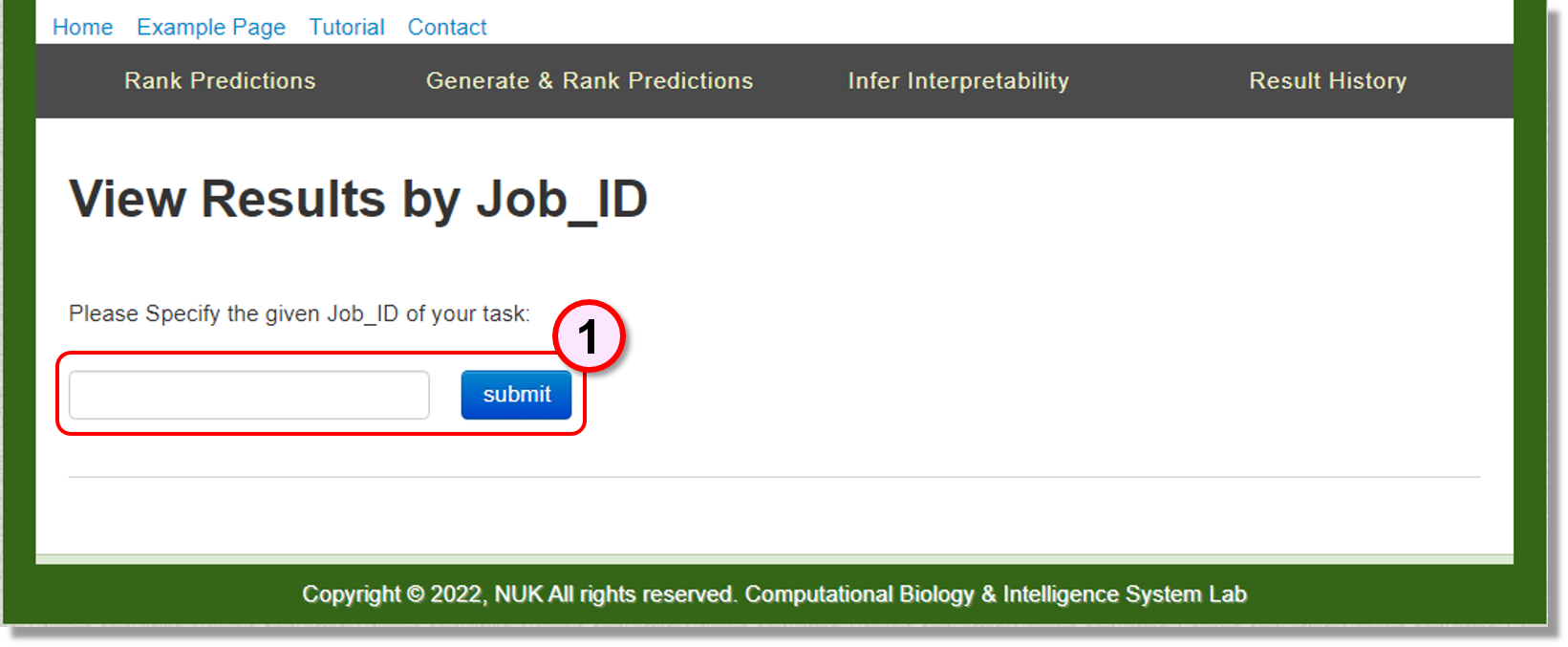
You can use the provided Job_id to find the identified RNA functionally interpretable structure ensemble in this page.
-
Input: Job_ID
Type in the given Job_ID to find the result.
Result page of "Rank Predictions"
The rankings of the list of predicted structures based on functional interpretability for the given sequence will be summarized in this page.
-
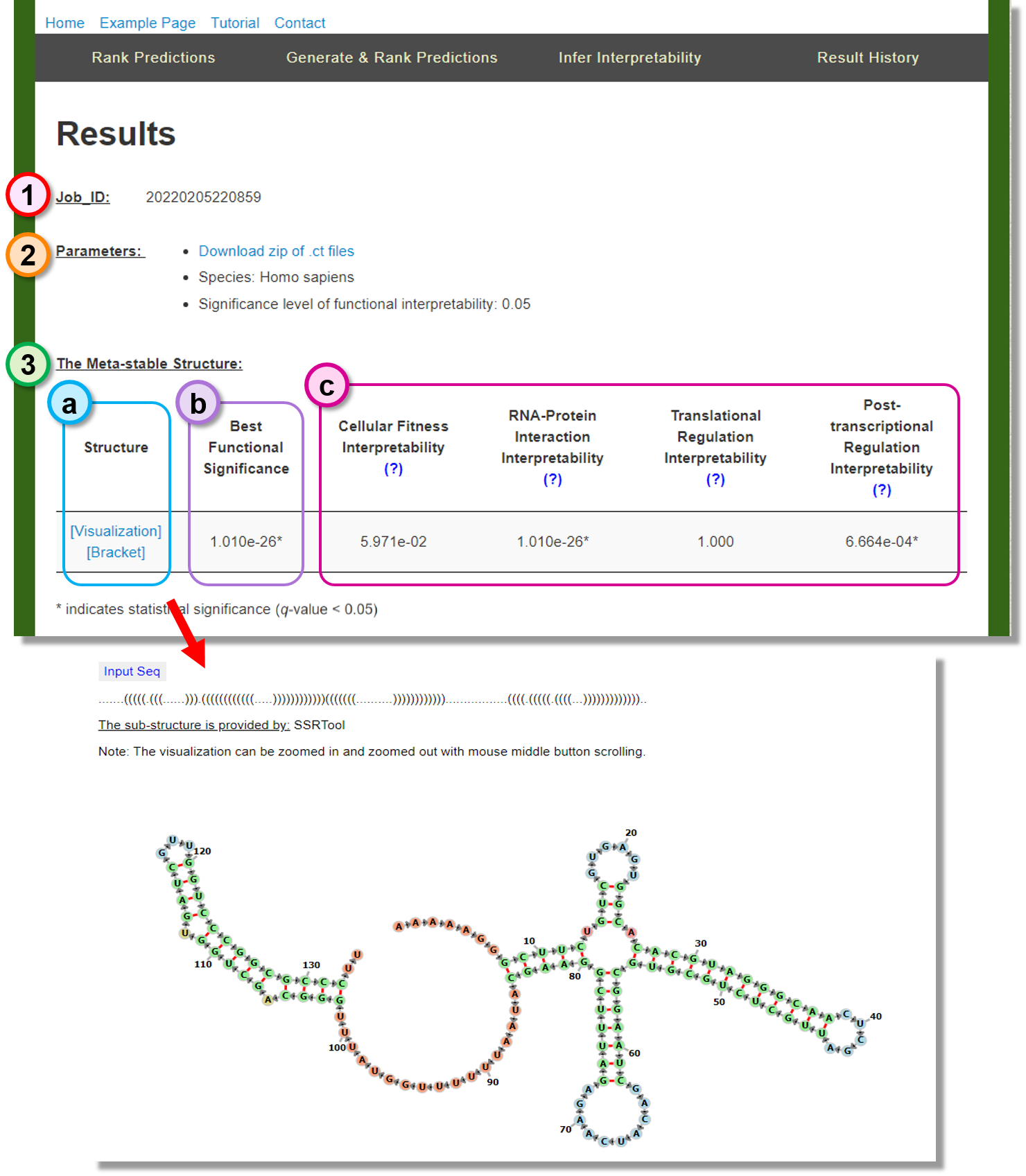
Job_ID
The web service Job_ID. -
Parameters
The user-specified algorithm parameters use to rank predictions. -
The Meta-stable Structure
The suggested most native-like candidate structure form the given conformation predictions.- (a) Structure: The meta-stable structure can be visualized or downloaded by clicking the provided link.
- (b) Best Functional Significance: The minimum significance in the functional interpretability metrics.
- (c) Significance metrics: The functionally interpretability test results of the meta-stable structure.
-
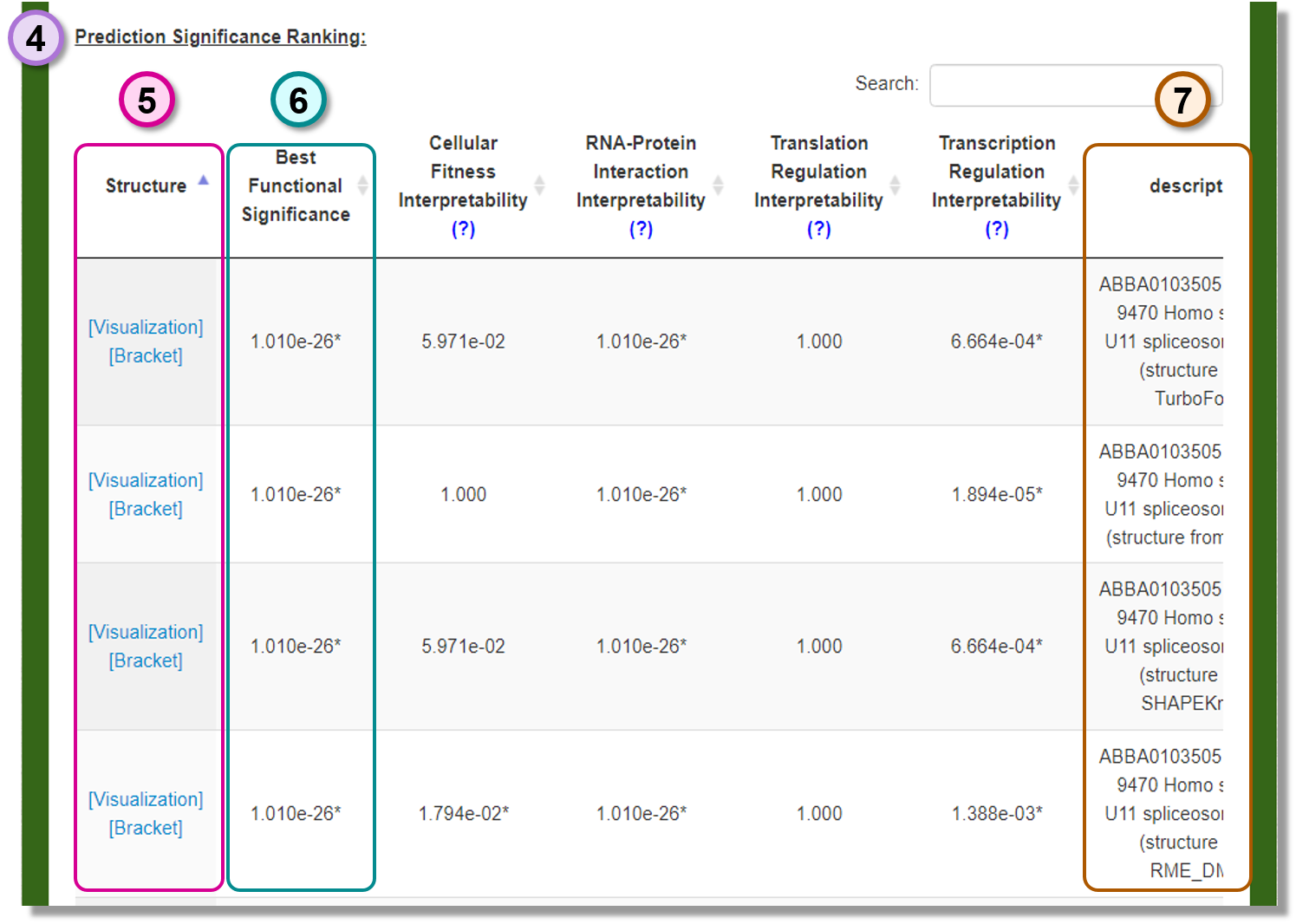
Prediction Significance Ranking
The functional rankings of the provided list of structure predictions. -
Structure
The predicted structure can be visualized or downloaded by clicking the provided link. -
Best Functional Significance
The minimum significance for the structure predictions. -
Description
The user specified information for the structure predictions.
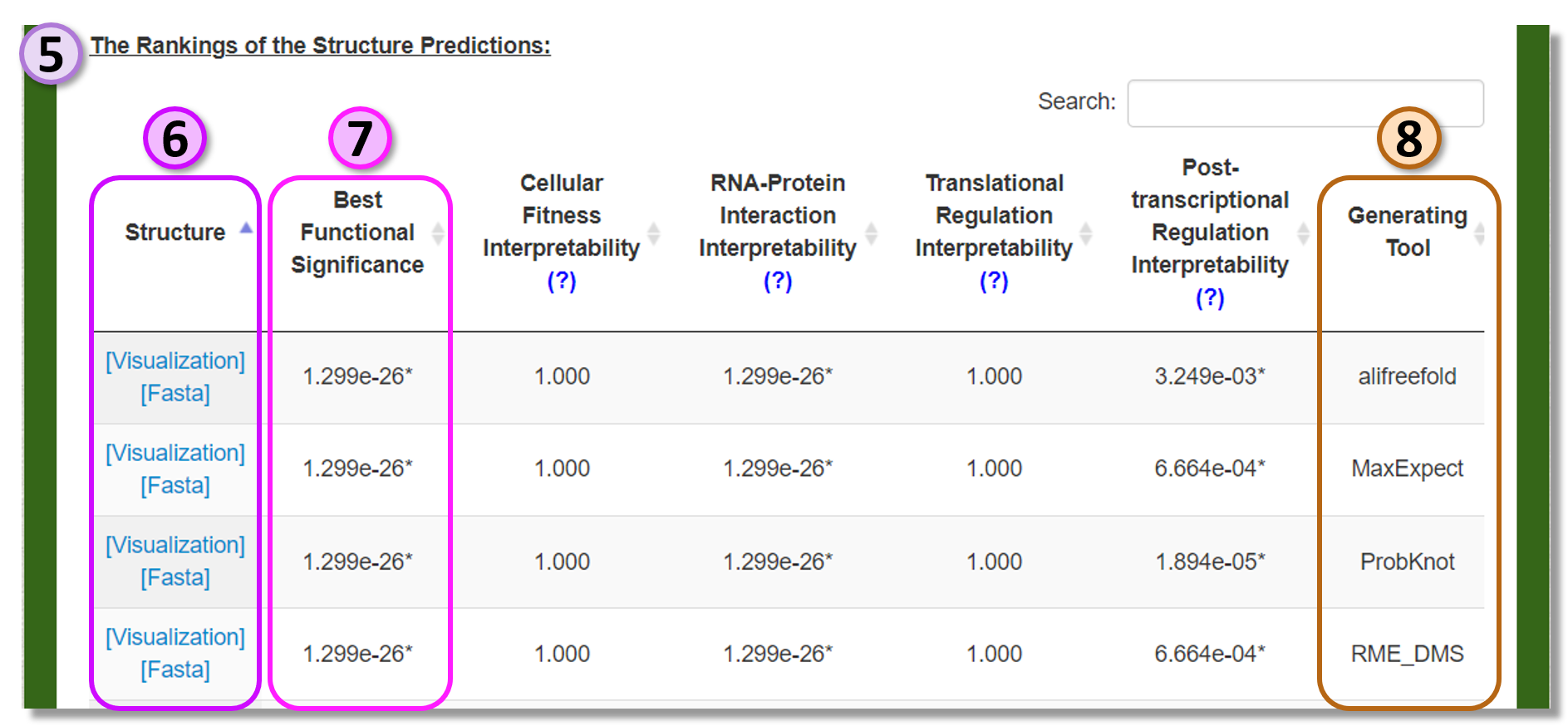
Result page of "Generate & Rank Predictions"
The generated structure predictions by the specified prediction tools and their rankings are summarized in this page.
-
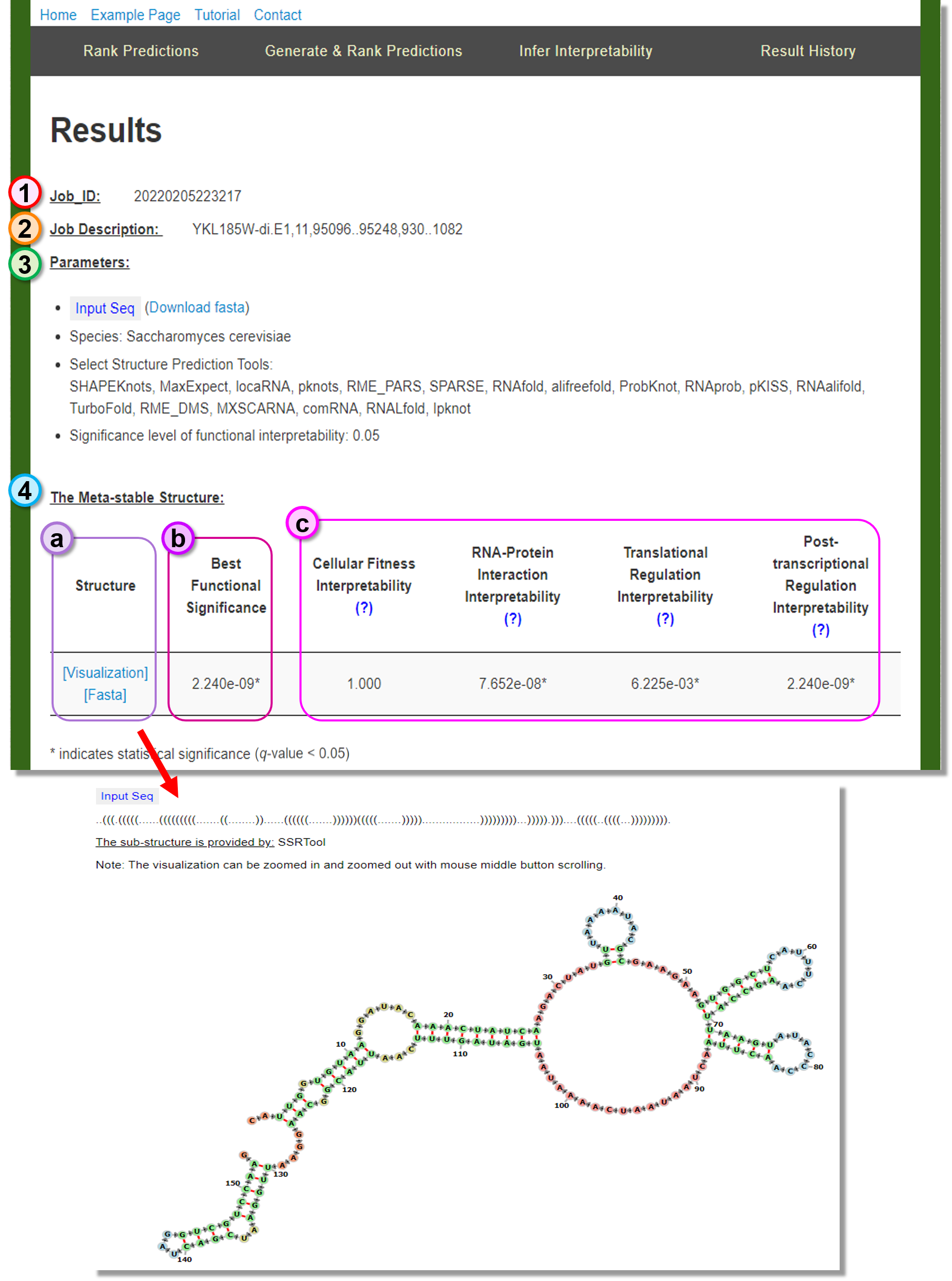
Job_ID
The web service Job_ID of. -
Job Description
The sequence information provided in the header of the given .fasta file. -
Parameters
The user-specified algorithm parameters. -
The Meta-stable Structure
The suggested most native-like candidate structure form the given conformation predictions.- (a) Structure: The meta-stable structure can be visualized or downloaded by clicking the provided link.
- (b) Best Functional Significance: The minimum significance in the functional interpretability metrics.
- (c) Significance metrics: The functionally interpretability test results of the meta-stable structure.
-
Structure
The predicted structure can be visualized or downloaded by clicking the provided link. -
Best Functional Significance
The minimum significance for the structure predictions. -
Generating tool
The prediction tool from which the sub-optimal structure was adopted.
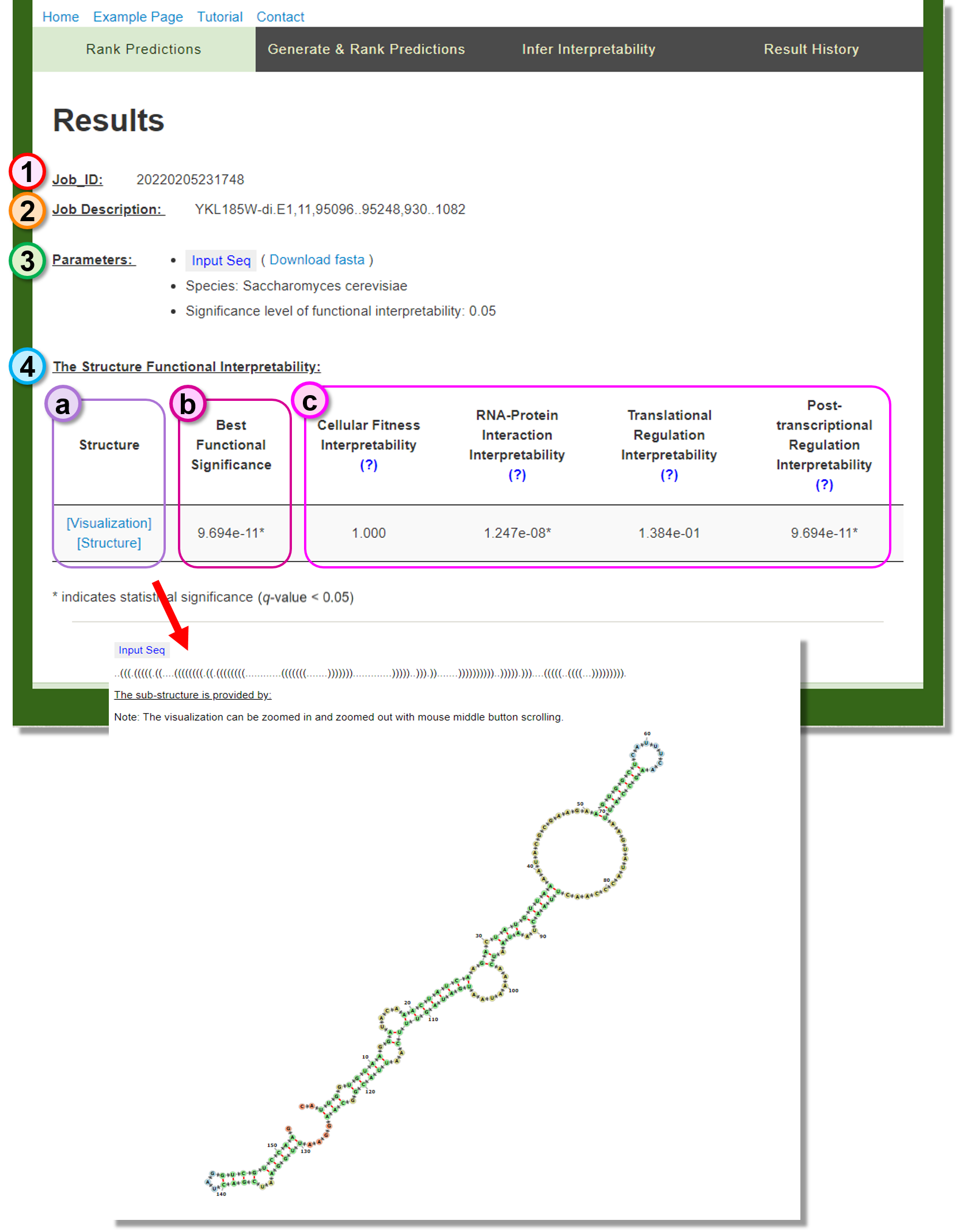
Result page of Infer Interpretability
Infer the functional interpretability for the given structure.
-
Job_ID
The web service Job_ID. -
Job Description
The sequence information provided in the header of the dot bracket file. -
Parameters
The user-specified algorithm parameters in identifying the functionally interpretable structure. -
The Structur Functional Interpretability
- (a) Structure: The meta-stable structure can be visualized or downloaded by clicking the provided link.
- (b) Best Functional Significance: The minimum significance in the functional interpretability metrics.
- (c) Significance metrics: The functionally interpretability test results of the meta-stable structure.
